注意
前往結尾下載完整的範例程式碼。或透過 JupyterLite 或 Binder 在您的瀏覽器中執行此範例
在斷裂球體上的流形學習方法#
在球形數據集上應用不同的 流形學習 技術。在這裡可以看到使用降維來獲得一些關於流形學習方法的直觀理解。關於數據集,極點從球體上切除,並且沿著側面切下一薄片。這使流形學習技術能夠在將其投影到二維的同時「展開」。
對於一個類似的範例,將這些方法應用於 S 曲線數據集,請參閱流形學習方法比較
請注意,MDS 的目的是找到數據的低維表示(此處為 2D),其中距離能很好地尊重原始高維空間中的距離,與其他流形學習演算法不同,它不尋求在低維空間中對數據進行各向同性表示。此處的流形問題與地球的平面地圖表示非常相似,就像 地圖投影 一樣
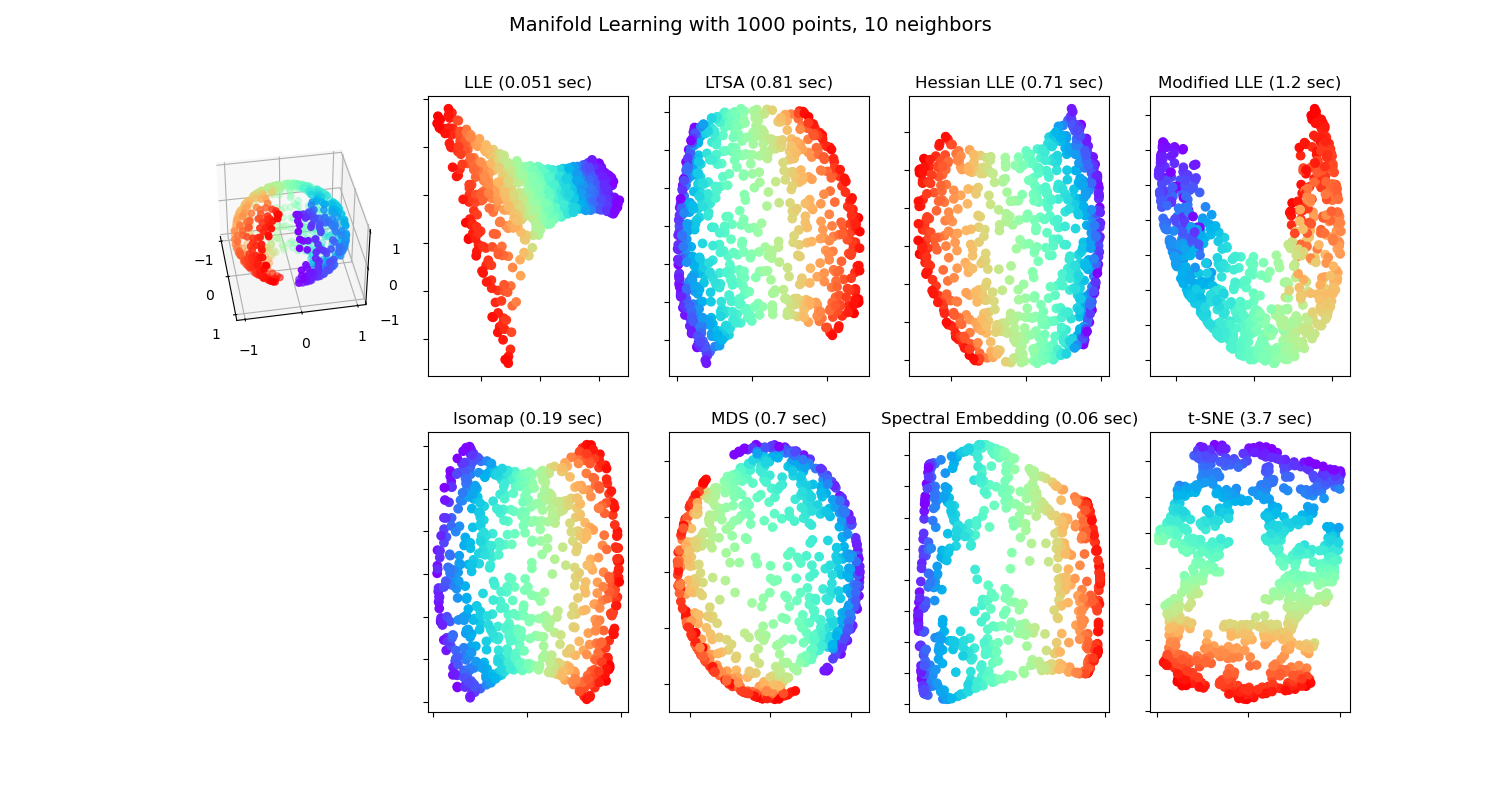
standard: 0.051 sec
ltsa: 0.81 sec
hessian: 0.71 sec
modified: 1.2 sec
ISO: 0.19 sec
MDS: 0.7 sec
Spectral Embedding: 0.06 sec
t-SNE: 3.7 sec
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
from time import time
import matplotlib.pyplot as plt
# Unused but required import for doing 3d projections with matplotlib < 3.2
import mpl_toolkits.mplot3d # noqa: F401
import numpy as np
from matplotlib.ticker import NullFormatter
from sklearn import manifold
from sklearn.utils import check_random_state
# Variables for manifold learning.
n_neighbors = 10
n_samples = 1000
# Create our sphere.
random_state = check_random_state(0)
p = random_state.rand(n_samples) * (2 * np.pi - 0.55)
t = random_state.rand(n_samples) * np.pi
# Sever the poles from the sphere.
indices = (t < (np.pi - (np.pi / 8))) & (t > ((np.pi / 8)))
colors = p[indices]
x, y, z = (
np.sin(t[indices]) * np.cos(p[indices]),
np.sin(t[indices]) * np.sin(p[indices]),
np.cos(t[indices]),
)
# Plot our dataset.
fig = plt.figure(figsize=(15, 8))
plt.suptitle(
"Manifold Learning with %i points, %i neighbors" % (1000, n_neighbors), fontsize=14
)
ax = fig.add_subplot(251, projection="3d")
ax.scatter(x, y, z, c=p[indices], cmap=plt.cm.rainbow)
ax.view_init(40, -10)
sphere_data = np.array([x, y, z]).T
# Perform Locally Linear Embedding Manifold learning
methods = ["standard", "ltsa", "hessian", "modified"]
labels = ["LLE", "LTSA", "Hessian LLE", "Modified LLE"]
for i, method in enumerate(methods):
t0 = time()
trans_data = (
manifold.LocallyLinearEmbedding(
n_neighbors=n_neighbors, n_components=2, method=method, random_state=42
)
.fit_transform(sphere_data)
.T
)
t1 = time()
print("%s: %.2g sec" % (methods[i], t1 - t0))
ax = fig.add_subplot(252 + i)
plt.scatter(trans_data[0], trans_data[1], c=colors, cmap=plt.cm.rainbow)
plt.title("%s (%.2g sec)" % (labels[i], t1 - t0))
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
plt.axis("tight")
# Perform Isomap Manifold learning.
t0 = time()
trans_data = (
manifold.Isomap(n_neighbors=n_neighbors, n_components=2)
.fit_transform(sphere_data)
.T
)
t1 = time()
print("%s: %.2g sec" % ("ISO", t1 - t0))
ax = fig.add_subplot(257)
plt.scatter(trans_data[0], trans_data[1], c=colors, cmap=plt.cm.rainbow)
plt.title("%s (%.2g sec)" % ("Isomap", t1 - t0))
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
plt.axis("tight")
# Perform Multi-dimensional scaling.
t0 = time()
mds = manifold.MDS(2, max_iter=100, n_init=1, random_state=42)
trans_data = mds.fit_transform(sphere_data).T
t1 = time()
print("MDS: %.2g sec" % (t1 - t0))
ax = fig.add_subplot(258)
plt.scatter(trans_data[0], trans_data[1], c=colors, cmap=plt.cm.rainbow)
plt.title("MDS (%.2g sec)" % (t1 - t0))
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
plt.axis("tight")
# Perform Spectral Embedding.
t0 = time()
se = manifold.SpectralEmbedding(
n_components=2, n_neighbors=n_neighbors, random_state=42
)
trans_data = se.fit_transform(sphere_data).T
t1 = time()
print("Spectral Embedding: %.2g sec" % (t1 - t0))
ax = fig.add_subplot(259)
plt.scatter(trans_data[0], trans_data[1], c=colors, cmap=plt.cm.rainbow)
plt.title("Spectral Embedding (%.2g sec)" % (t1 - t0))
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
plt.axis("tight")
# Perform t-distributed stochastic neighbor embedding.
t0 = time()
tsne = manifold.TSNE(n_components=2, random_state=0)
trans_data = tsne.fit_transform(sphere_data).T
t1 = time()
print("t-SNE: %.2g sec" % (t1 - t0))
ax = fig.add_subplot(2, 5, 10)
plt.scatter(trans_data[0], trans_data[1], c=colors, cmap=plt.cm.rainbow)
plt.title("t-SNE (%.2g sec)" % (t1 - t0))
ax.xaxis.set_major_formatter(NullFormatter())
ax.yaxis.set_major_formatter(NullFormatter())
plt.axis("tight")
plt.show()
腳本總執行時間:(0 分鐘 7.884 秒)
相關範例