注意
前往結尾以下載完整的範例程式碼,或透過 JupyterLite 或 Binder 在您的瀏覽器中執行此範例
階層式分群:結構化 vs 非結構化 Ward#
範例建立瑞士捲資料集並在其位置上執行階層式分群。
如需更多資訊,請參閱階層式分群。
第一步,在沒有結構連通性限制的情況下執行階層式分群,並且僅基於距離,而在第二步中,分群被限制為 k-Nearest Neighbors 圖:這是一個具有結構先驗的階層式分群。
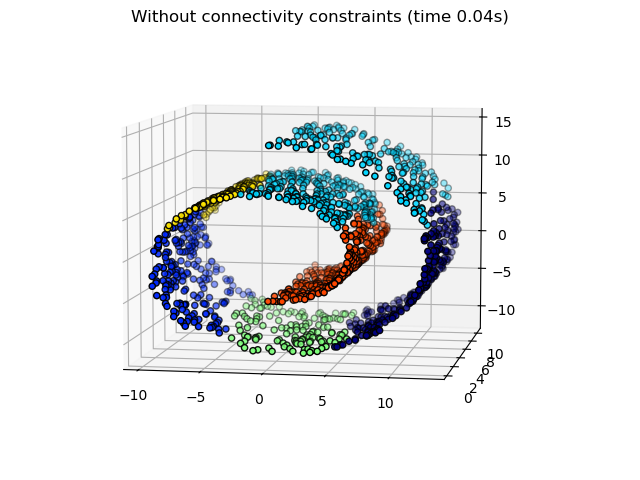
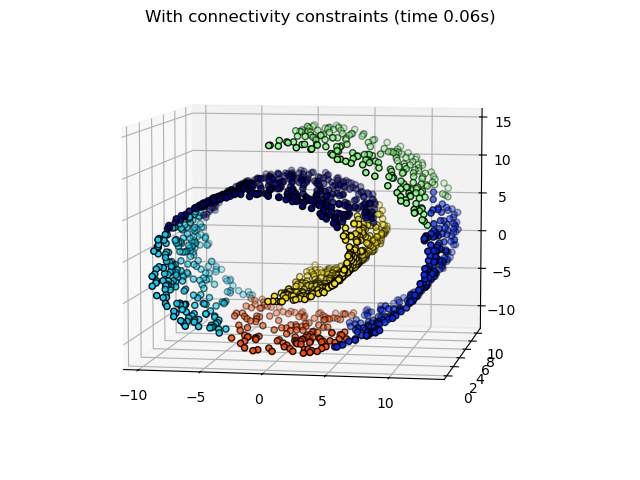
一些在沒有連通性限制的情況下學習到的分群不尊重瑞士捲的結構,並且延伸到多個流形褶皺。相反地,當反對連通性限制時,分群會形成瑞士捲的良好分割。
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import time as time
# The following import is required
# for 3D projection to work with matplotlib < 3.2
import mpl_toolkits.mplot3d # noqa: F401
import numpy as np
產生資料#
我們首先產生瑞士捲資料集。
from sklearn.datasets import make_swiss_roll
n_samples = 1500
noise = 0.05
X, _ = make_swiss_roll(n_samples, noise=noise)
# Make it thinner
X[:, 1] *= 0.5
計算分群#
我們執行 AgglomerativeClustering,它屬於沒有任何連通性限制的階層式分群。
from sklearn.cluster import AgglomerativeClustering
print("Compute unstructured hierarchical clustering...")
st = time.time()
ward = AgglomerativeClustering(n_clusters=6, linkage="ward").fit(X)
elapsed_time = time.time() - st
label = ward.labels_
print(f"Elapsed time: {elapsed_time:.2f}s")
print(f"Number of points: {label.size}")
Compute unstructured hierarchical clustering...
Elapsed time: 0.04s
Number of points: 1500
繪製結果#
繪製非結構化的階層式分群。
import matplotlib.pyplot as plt
fig1 = plt.figure()
ax1 = fig1.add_subplot(111, projection="3d", elev=7, azim=-80)
ax1.set_position([0, 0, 0.95, 1])
for l in np.unique(label):
ax1.scatter(
X[label == l, 0],
X[label == l, 1],
X[label == l, 2],
color=plt.cm.jet(float(l) / np.max(label + 1)),
s=20,
edgecolor="k",
)
_ = fig1.suptitle(f"Without connectivity constraints (time {elapsed_time:.2f}s)")
我們正在定義具有 10 個鄰居的 k-Nearest Neighbors#
from sklearn.neighbors import kneighbors_graph
connectivity = kneighbors_graph(X, n_neighbors=10, include_self=False)
計算分群#
我們再次執行具有連通性限制的 AgglomerativeClustering。
print("Compute structured hierarchical clustering...")
st = time.time()
ward = AgglomerativeClustering(
n_clusters=6, connectivity=connectivity, linkage="ward"
).fit(X)
elapsed_time = time.time() - st
label = ward.labels_
print(f"Elapsed time: {elapsed_time:.2f}s")
print(f"Number of points: {label.size}")
Compute structured hierarchical clustering...
Elapsed time: 0.06s
Number of points: 1500
繪製結果#
繪製結構化的階層式分群。
fig2 = plt.figure()
ax2 = fig2.add_subplot(121, projection="3d", elev=7, azim=-80)
ax2.set_position([0, 0, 0.95, 1])
for l in np.unique(label):
ax2.scatter(
X[label == l, 0],
X[label == l, 1],
X[label == l, 2],
color=plt.cm.jet(float(l) / np.max(label + 1)),
s=20,
edgecolor="k",
)
fig2.suptitle(f"With connectivity constraints (time {elapsed_time:.2f}s)")
plt.show()
腳本的總執行時間:(0 分鐘 0.368 秒)
相關範例