注意
前往結尾 下載完整的範例程式碼。或通過 JupyterLite 或 Binder 在您的瀏覽器中執行此範例
在建立估算器之前填補缺失值#
缺失值可以使用基本 SimpleImputer 替換為平均值、中位數或最常見的值。
在此範例中,我們將研究不同的填補技術
以常數值 0 填補
以每個特徵的平均值填補,並結合缺失指標輔助變數
k 最近鄰填補
迭代填補
我們將使用兩個資料集:糖尿病資料集,其中包含從糖尿病患者收集的 10 個特徵變數,旨在預測疾病進展;以及加州住房資料集,其目標是加州地區的房屋中位數價值。
由於這些資料集都沒有缺失值,我們將移除一些值以建立具有人工缺失資料的新版本。然後,比較 RandomForestRegressor 在完整原始資料集上的效能與在改變後的資料集上的效能,其中使用不同技術填補了人工缺失值。
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
下載資料並建立缺失值集#
首先,我們下載兩個資料集。糖尿病資料集隨 scikit-learn 一起提供。它有 442 個條目,每個條目有 10 個特徵。加州住房資料集較大,有 20640 個條目和 8 個特徵。它需要下載。為了加快計算速度,我們只會使用前 400 個條目,但請隨意使用整個資料集。
import numpy as np
from sklearn.datasets import fetch_california_housing, load_diabetes
rng = np.random.RandomState(42)
X_diabetes, y_diabetes = load_diabetes(return_X_y=True)
X_california, y_california = fetch_california_housing(return_X_y=True)
X_california = X_california[:300]
y_california = y_california[:300]
X_diabetes = X_diabetes[:300]
y_diabetes = y_diabetes[:300]
def add_missing_values(X_full, y_full):
n_samples, n_features = X_full.shape
# Add missing values in 75% of the lines
missing_rate = 0.75
n_missing_samples = int(n_samples * missing_rate)
missing_samples = np.zeros(n_samples, dtype=bool)
missing_samples[:n_missing_samples] = True
rng.shuffle(missing_samples)
missing_features = rng.randint(0, n_features, n_missing_samples)
X_missing = X_full.copy()
X_missing[missing_samples, missing_features] = np.nan
y_missing = y_full.copy()
return X_missing, y_missing
X_miss_california, y_miss_california = add_missing_values(X_california, y_california)
X_miss_diabetes, y_miss_diabetes = add_missing_values(X_diabetes, y_diabetes)
填補缺失資料並評分#
現在我們將編寫一個函數,該函數將對不同填補的資料上的結果進行評分。讓我們分別看看每個填補器
rng = np.random.RandomState(0)
from sklearn.ensemble import RandomForestRegressor
# To use the experimental IterativeImputer, we need to explicitly ask for it:
from sklearn.experimental import enable_iterative_imputer # noqa
from sklearn.impute import IterativeImputer, KNNImputer, SimpleImputer
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import make_pipeline
N_SPLITS = 4
regressor = RandomForestRegressor(random_state=0)
缺失資訊#
除了填補缺失值之外,填補器還具有一個 add_indicator 參數,用於標記缺失的值,這些值可能攜帶一些資訊。
def get_scores_for_imputer(imputer, X_missing, y_missing):
estimator = make_pipeline(imputer, regressor)
impute_scores = cross_val_score(
estimator, X_missing, y_missing, scoring="neg_mean_squared_error", cv=N_SPLITS
)
return impute_scores
x_labels = []
mses_california = np.zeros(5)
stds_california = np.zeros(5)
mses_diabetes = np.zeros(5)
stds_diabetes = np.zeros(5)
估計分數#
首先,我們要估計原始資料的分數
def get_full_score(X_full, y_full):
full_scores = cross_val_score(
regressor, X_full, y_full, scoring="neg_mean_squared_error", cv=N_SPLITS
)
return full_scores.mean(), full_scores.std()
mses_california[0], stds_california[0] = get_full_score(X_california, y_california)
mses_diabetes[0], stds_diabetes[0] = get_full_score(X_diabetes, y_diabetes)
x_labels.append("Full data")
將缺失值替換為 0#
現在,我們將估計缺失值被替換為 0 的資料上的分數
def get_impute_zero_score(X_missing, y_missing):
imputer = SimpleImputer(
missing_values=np.nan, add_indicator=True, strategy="constant", fill_value=0
)
zero_impute_scores = get_scores_for_imputer(imputer, X_missing, y_missing)
return zero_impute_scores.mean(), zero_impute_scores.std()
mses_california[1], stds_california[1] = get_impute_zero_score(
X_miss_california, y_miss_california
)
mses_diabetes[1], stds_diabetes[1] = get_impute_zero_score(
X_miss_diabetes, y_miss_diabetes
)
x_labels.append("Zero imputation")
缺失值的 kNN 填補#
KNNImputer 使用所需數量的最近鄰的加權或未加權平均值來填補缺失值。
def get_impute_knn_score(X_missing, y_missing):
imputer = KNNImputer(missing_values=np.nan, add_indicator=True)
knn_impute_scores = get_scores_for_imputer(imputer, X_missing, y_missing)
return knn_impute_scores.mean(), knn_impute_scores.std()
mses_california[2], stds_california[2] = get_impute_knn_score(
X_miss_california, y_miss_california
)
mses_diabetes[2], stds_diabetes[2] = get_impute_knn_score(
X_miss_diabetes, y_miss_diabetes
)
x_labels.append("KNN Imputation")
以平均值填補缺失值#
def get_impute_mean(X_missing, y_missing):
imputer = SimpleImputer(missing_values=np.nan, strategy="mean", add_indicator=True)
mean_impute_scores = get_scores_for_imputer(imputer, X_missing, y_missing)
return mean_impute_scores.mean(), mean_impute_scores.std()
mses_california[3], stds_california[3] = get_impute_mean(
X_miss_california, y_miss_california
)
mses_diabetes[3], stds_diabetes[3] = get_impute_mean(X_miss_diabetes, y_miss_diabetes)
x_labels.append("Mean Imputation")
缺失值的迭代填補#
另一個選項是 IterativeImputer。這使用循環線性迴歸,依次將每個具有缺失值的特徵建模為其他特徵的函數。實施的版本假定高斯(輸出)變數。如果您的特徵顯然是非常態的,請考慮轉換它們以使其看起來更常態,以潛在提高效能。
def get_impute_iterative(X_missing, y_missing):
imputer = IterativeImputer(
missing_values=np.nan,
add_indicator=True,
random_state=0,
n_nearest_features=3,
max_iter=1,
sample_posterior=True,
)
iterative_impute_scores = get_scores_for_imputer(imputer, X_missing, y_missing)
return iterative_impute_scores.mean(), iterative_impute_scores.std()
mses_california[4], stds_california[4] = get_impute_iterative(
X_miss_california, y_miss_california
)
mses_diabetes[4], stds_diabetes[4] = get_impute_iterative(
X_miss_diabetes, y_miss_diabetes
)
x_labels.append("Iterative Imputation")
mses_diabetes = mses_diabetes * -1
mses_california = mses_california * -1
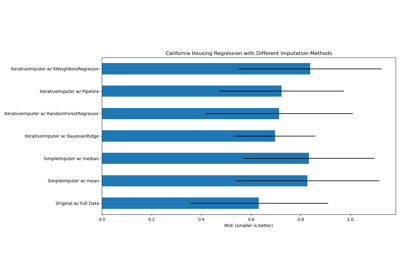
繪製結果#
最後,我們將視覺化分數
import matplotlib.pyplot as plt
n_bars = len(mses_diabetes)
xval = np.arange(n_bars)
colors = ["r", "g", "b", "orange", "black"]
# plot diabetes results
plt.figure(figsize=(12, 6))
ax1 = plt.subplot(121)
for j in xval:
ax1.barh(
j,
mses_diabetes[j],
xerr=stds_diabetes[j],
color=colors[j],
alpha=0.6,
align="center",
)
ax1.set_title("Imputation Techniques with Diabetes Data")
ax1.set_xlim(left=np.min(mses_diabetes) * 0.9, right=np.max(mses_diabetes) * 1.1)
ax1.set_yticks(xval)
ax1.set_xlabel("MSE")
ax1.invert_yaxis()
ax1.set_yticklabels(x_labels)
# plot california dataset results
ax2 = plt.subplot(122)
for j in xval:
ax2.barh(
j,
mses_california[j],
xerr=stds_california[j],
color=colors[j],
alpha=0.6,
align="center",
)
ax2.set_title("Imputation Techniques with California Data")
ax2.set_yticks(xval)
ax2.set_xlabel("MSE")
ax2.invert_yaxis()
ax2.set_yticklabels([""] * n_bars)
plt.show()
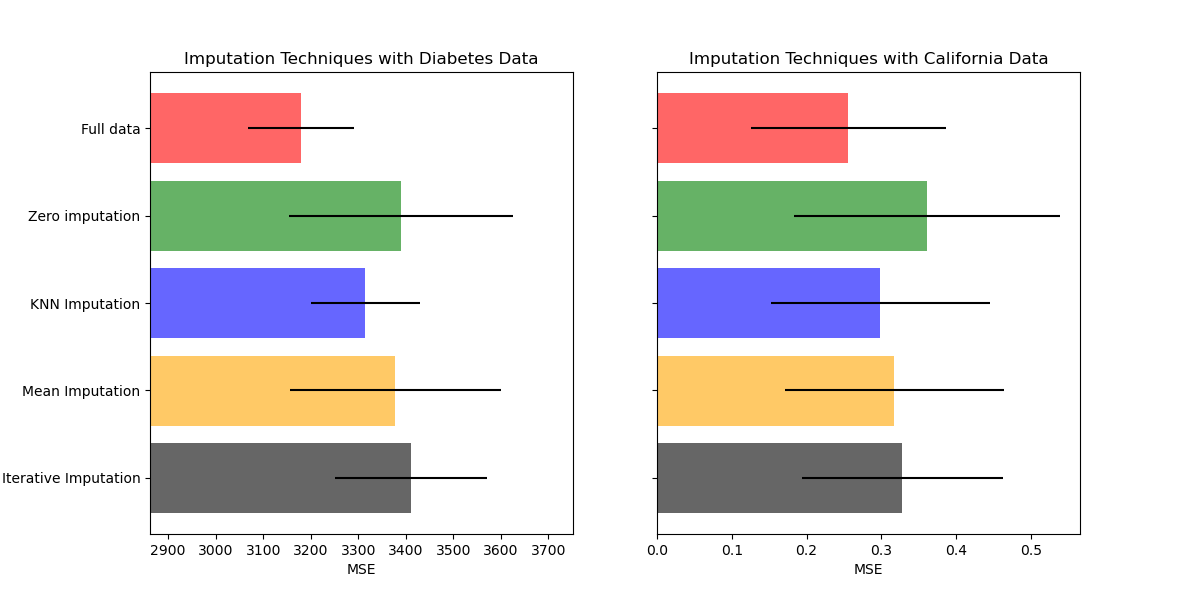
您也可以嘗試不同的技術。例如,對於具有高幅度變數(也稱為「長尾」)的資料,中位數是更穩健的估算器,這些變數可能會主導結果。
腳本的總執行時間:(0 分鐘 11.125 秒)
相關範例